| Category | Coursework | Subject | Computer Science |
|---|---|---|---|
| University | University of Westminster | Module Title | 7BUIS008W.1 Data Mining & Machine Learning |
Students are expected to critically engage in effectively applying and evaluating novel data mining and machine learning techniques for a specific problem domain and definitely reflect on the knowledge of how different data mining and machine learning algorithms perform in terms of biases for a given problem domain. Students are expected to methodically analyse the output of the data mining tasks and machine learning algorithms by drawing technically appropriate and sound conclusions resulting from the application of data mining and machine learning algorithms to the given problem.
This assignment contributes towards the following Learning Outcomes (LOs):
A) The Domain
The deployment of machine learning modelling in this coursework aims to tackle a real-world tool by developing effective early screening machine learning models for breast cancer mortality and survival prediction to help doctors enhance their treatment planning and management.
Cancer is a disease in which cells in the body grow out of control. Breast cancer is a disease in which abnormal breast cells grow out of control and form tumours. If left unchecked, the tumours can spread throughout the body and become fatal. Breast cancer cells begin inside the milk ducts and/or the milk- producing lobules of the breast.
In females in the UK, breast cancer is the 2nd most common cause of cancer death, with around 11,400 deaths every year (2017-2019). In males in the UK, breast cancer is not among the 20 most common causes of cancer death, with around 85 deaths every year (2017-2019).
The tests and scans the patient have to diagnose breast cancer give information about:
The tests the patient has also give information about the type of breast cancer they have. The information below is an overview of the TNM staging for all types of cancer.
The stage of cancer tells the patient how big it is and how far it has spread. It helps the doctor decide the best treatment for the
patient. There are different systems used in the UK to stage breast cancer. Stage 1 is part of the number staging system. Doctors may also use the TNM staging system.
Staging for breast cancer is very complex. Many different factors are considered before doctors can confirm the patient’s final stage.
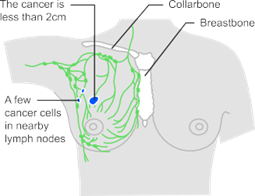
Fig.1 Illustration of stage 1 breast cancer
Stage 2 breast cancer means that the cancer is either in the breast or in the nearby lymph nodes or both. It is an early-stage breast cancer.
Stage 2 is part of the number staging system. Doctors may also use the TNM staging system.
Stage 2 can be divided into 2A and 2B. Opposite is a simplified description of stage 2A and 2B breast cancer (see Figure 2).
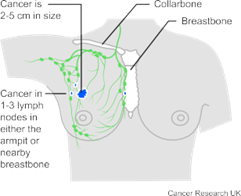
Fig.2 Illustration of stage 2 breast cancer
Stage 3 means that the cancer has spread from the breast to the lymph nodes close to the breast, to the skin of the breast or to the chest wall. It is also called locally advanced breast cancer. Stage 3 is part of the number staging system. Doctors may also use the TNM staging system.
Stage 3 can be divided into 3A, 3B and 3C. opposite is a simplified description of stage 3A, 3B and 3C breast cancer (see Figure 3).
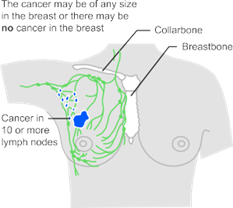
Fig.3 Illustration of stage 3 breast cancer
Stage 4 breast cancer has spread to another part of the body (see Figure 4). It is also called advanced cancer or secondary breast cancer. The aim of treatment is to control the cancer and any symptoms. Treatment depends on a number of factors.
In stage 4 breast cancer:
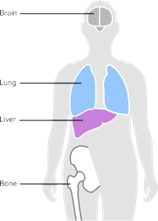
Fig.4 Illustration of stage 4 breast cancer
Hormonal Treatment
Hormone therapy is a common treatment for secondary breast cancer. It can often shrink and control the cancer wherever it is in the body. It works well if the cancer cells have particular proteins called hormone receptors, estrogen receptor, progesterone receptor.
If one hormone therapy stops working so well, the doctor might suggest you try a different one.
Other Treatment
Doctor will take many different factors into account when deciding which treatment is best for the patient. These include:
If your cancer doesn't have hormone receptors or has spread to the liver or lungs, the doctor might suggest Chemotherapy. Radiotherapy might be recommended if the cancer has spread to the bones or the skin near the breast. Targeted and immunotherapy drugs might be recommended for secondary breast cancer.
C) The Domain Problem
The importance of predicting mortality, short- and long-term survival of patients with cancer may improve their care. Prior predictive models either use data with limited availability or predict the outcome of only 1 type of cancer. In this case breast cancer.
D) Your Role as A Data Scientist
You are hired as a data scientist to work alongside a team of doctors to
1- Build predictive machine-learning models for breast cancer mortality status.
2- Build predictive machine-learning models to estimate patient’s survival period.
The team of doctors provided you with historical records of breast cancer patients and had their mortality status. Also, obtained the number of months they survived.
The doctors rely on your work to answer the following two research question on the dataset; the key objective is to create a new, predictive tool powered by a machine learning model to assist doctors in enhancing their treatment planning and cancer care. The Research Questions are:
a) Does machine learning have the potential to assist doctors to predict those who would survive breast cancer or not?
b) For patients who would not survive cancer, can machine learning offer a reliable estimate of their survival period?
E) Your Dataset
This dataset of breast cancer patients was obtained from the 2017 November update of the SEER Program of the NCI, which provides information on population-based cancer statistics. The dataset contains the following attributes:
As a data scientist, you are a logician, a mathematician, a technician, and an analyst, and you need doctors to understand your analyses. Doctors are usually busy individuals, and they don’t have all the time in the world. One essential skill that you must adhere to is to be concise and straight to the point. Focus on the answers needed for each task, and provide just enough words for the answer only. There is no need to provide lengthy descriptions of algorithms and methods unless you are asked to do.
Also, they are only interested in assessing your interpretation of the modelling results, so you MUST NOT paste any Python code in this report unless specifically asked to so. You will receive a separate link to submit your code as a Python notebook file (mandatory). ipynb extension. Your data mining tasks will be aligned with the popular CRISP-DM methodology phases but without the deployment phase (see Figure 5).
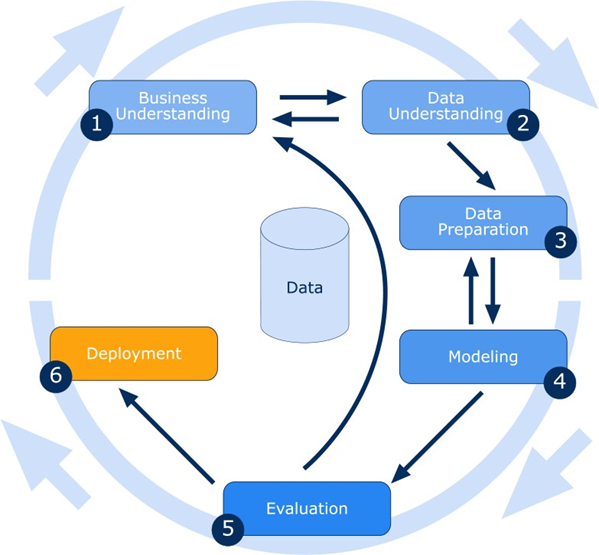
Fig.2 CRISP-DM Phases
Does machine learning have the potential to assist doctors to predict those who would survive breast cancer or not Task (1) – Domain Understanding:
Classification [Total 6 Marks] The doctors decided that classification modelling is required. Indicate in the table below for each of the listed variables in your data which ones you should RETAIN and can be included in the classification modelling of Breast Cancer Mortality (Alive vs. Dead) and the variables you should DROP (REMOVE). Justify your decision logically and/or by research (include in-text citation)
|
Variable Name |
Retain or Drop |
Brief justification for retention or dropping |
|
Patient ID |
|
|
|
Month of Birth |
|
|
|
Age |
|
|
|
Sex |
|
|
|
Race |
|
|
|
Marital Status |
|
|
|
Occupation Code |
|
|
|
Adopted Status |
|
|
|
T Stage |
|
|
|
N Stage |
|
|
|
6th Stage |
|
|
|
Differentiate |
|
|
|
Grade |
|
|
|
A Stage |
|
|
|
Tumour Size |
|
|
|
Estrogen Status |
|
|
|
Progesterone Status |
|
|
|
Regional Node Examined |
|
|
|
Regional Node Positive |
|
|
|
Survival Months |
|
|
|
Mortality Status |
|
|
From your Python notebook, for your RETAINED input variables and your class “target” variable, produce a basic statistical description and variable scale type. Plot the distribution of your target variable. (Paste screenshots of code OUTPUTS ONLY for evidence).
a) Investigate any issues in your retained dataset and the possible variables. Based on the issues you find in your data, suggest a suitable possible method to mitigate each of these issues and provide your justification for using each method. Use the table below to organise your findings, add more rows if needed:
|
Variable Name |
Issue description |
Proposed mitigation |
Justification for used mitigation |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
⋮ |
⋮ |
⋮ |
⋮ |
b) With the aid of Python packages and a notebook, implement your suggested mitigations of issues in Task (3.a) ,and show evidence of implementing your suggested solutions to the problems you identified for your dataset in (Task 3. a). (Use screenshots of code OUTPUTS ONLY). Indicate and annotate in your screenshots which issue was resolved from each screenshot provided. Show screenshots of code outputs before and after implementing your solution.
a) From the classification algorithms which you learned in the module, four different algorithms were selected: Logistic Regression (LR), K Nearest Neighbour (KNN) and Naïve Bayes (NB). These algorithms are a mix of parametric and non-parametric algorithms. List down the type of each algorithm (parametric vs non-parametric), name any learnable parameters, and list any possible hyperparameters for each algorithm which you may want to consider tuning. Note the Python package and module for importing each algorithm. Again, organise your answer in a table as before. See below:
|
Algorithm Name |
Algorithm Type |
Learnable Parameters |
Some Possible Hyperparameters |
Imported Python package to use the algorithm |
|
NB |
|
|
|
|
|
LR |
|
|
|
|
|
KNN (N=?) |
|
|
|
|
b) With the aid of the Python packages, use the training–test split approach with your retained applicable categorical input features only and the class output feature to build your predictive classification models.
Your healthcare professionals provided the following success criteria to guide you when evaluating your models.
“When evaluating your model's performance, which addresses your first research question (a). The model is expected to misclassify subjects. Thus, the model should aim to predict the “Dead” mortality status of subjects for as many as possible to increase the urgency in treatment planning. However, the model should demonstrate that its high “Death” mortality prediction rate is mainly due to a larger portion of correctly detected (predicted) subjects who belong to the Mortality Status “Dead” class.”
a) With the aid of Python packages, paste the test confusion matrix for each trained model as screenshots from the output of your Python code.
b) Five different classification evaluation metrics are noted. Paste each model’s test performance results. State which evaluation metric/metrics to “USE or “NOT USE” to closely interpret the above success criteria. For justification, explain how closely your choice of “USE” or “DO NOT USE” for a metric interprets the given success criteria. With the aid of Python packages, document the TEST SCORES for each built model
|
Metrics |
USE or DO NOT USE |
Justification in relation to the success criteria |
Model Name |
Test Score |
|
Accuracy |
|
|
NB |
|
|
LR |
|
|||
|
KNN (K=?) |
|
|||
|
Recall |
|
|
NB |
|
|
LR |
|
|||
|
KNN (K=?) |
|
|||
|
Precision |
|
|
NB |
|
|
LR |
|
|||
|
KNN (K=?) |
|
|||
|
F-Score |
|
|
NB |
|
|
LR |
|
|||
|
KNN (K=?) |
|
|||
|
AUC-ROC |
|
|
NB |
|
|
LR |
|
|||
|
KNN (K=?) |
|
c) Suggest a single best classification model based on the ‘USED’ performance metrics scores you identified in (Task 5. b). Briefly describe how well your best model satisfies the needs of your healthcare professionals.
d) Investigated with evidence to establish whether your selected best model is good fit, underfit or overfit.
e) To enhance your selected best model/s performance, tune some of its possible hyperparameters, which you indicated in (Task 4. a) for that specific algorithm. With the aid of Python packages, Re-train the algorithm again with GridSearchCV [5 marks]
i. Indicate the number of cross-validation K folds used.
ii. For the newly tuned model, document the estimated best hyperparameters,
iii. Present the test confusion matrix for the best models before and after tuning.
iv. Calculate and document the new score/s of the “USED” performance metric/s of your choice to interpret the success criteria identified in (Task 5.b) before and after tuning.
v. Use your observations to comment on whether the tuning of hyperparameters of your best model improved its positive predictive ability in line with the success criteria.
f) Based on your best model, draft an answer for the research question, criticise your best-performing model, and state any limitations you may have identified. Research and try to explain why your selected algorithm overtook all other models in no more than 100 words. State any ethical issues your model may raise if used to screen for breast cancer mortality.
The doctors decided that regression modelling is required. Using python functions, show the dimensions of your data subset that you will RETAIN for this regression modelling problem. Using python functions, list the names of the features that you intend to use for modelling from Table.1.
From your Python notebook, Plot the distribution for your RETAINED input variables and your “target” variable, (Paste screenshots of code OUTPUTS ONLY “the plots” for evidence).
a) By looking at the dataset establish whether there is a need for scaling your dataset attributes. Explain with evidence from your python code output the reasoning behind your recommendation.
b) In general, when applying scaling to any dataset for regression modelling, would scale the input features only, the target feature only, or all features? In less than 150 words, briefly justify your answer and include in-text citation where appropriate.
a) From the regression algorithms which you learned in the module, doctors decided on the use of a Decision Tree Regression (DT) algorithm. In less than 50 words, explain the added benefit of using a DT regressor to this healthcare prediction problem.
b) With the aid of the Python packages, you will use training – test split of 80:20 to build and test two DT regression models, Model 1 & Model 2. The first DT model with numeric features only and the second model using all your retained features:
i. From your python notebook, insert in your report, provide as evidence the code line from your source code that ensures reproducibility of your training - test sampling.
ii. Using python packages, show from your code output the dimensions of your training and test subsets used for each model. List the subset of features names used for Model 1 and Model 2.
Your healthcare professionals provided the following success criteria to guide you when evaluating your models. “When evaluating both models’ performances which addresses your research question (b), the model is expected to make some errors in estimating the survival months. However, the selected model out of the two built models should have input features that are better at explaining the recorded values of survival months.”
a) Four different regression evaluation metrics are noted. State which evaluation metric/metrics to USE or NOT USE to interpret the above success criteria closely. Justify your choice of USE or DO NOT USE. With the aid of Python packages, document the TEST SCORES for each built model.
|
Metrics |
USE or DO NOT USE |
Justification in relation to the success criteria |
Model Name |
Test Score |
|
MSE |
|
|
DT (Numeric Features Only) |
|
|
DT (All Features Only) |
|
|||
|
MAE |
|
|
DT (Numeric Features Only) |
|
|
DT (All Features Only) |
|
|||
|
R-Square |
|
|
DT (Numeric Features Only) |
|
|
DT (All Features Only) |
|
b) Describe any caveats to your selected performance metric assessing the ability of your model meeting the success criteria.
c) Suggest a single best regression model (Model 1 or Model 2) based on the ‘USED’ performance metrics scores you identified in (Task 5. b). Briefly describe how well your selected best model satisfies the needs of the healthcare professionals.
d) Health care professionals aim to explain your best model’s decision of estimated survival months to the patient. Therefore, rebuild your best model while performing pre-pruning (4 levels limit) to ease the interpretation of your best model’s decision. Plot the pruned tree and paste it here from your python notebook results. Describe with evidence if there were any performance advantages or disadvantages of pruning your best tree model.
e) Using your pruned model, predict the survival months for breast cancer patient B002565 whose attributes values are the following:
|
Variable Name |
Value |
|
Patient ID |
B002565 |
|
Month of Birth |
July |
|
Age |
56 Years old |
|
Sex |
Female |
|
Race |
White |
|
Marital Status |
Single |
|
Occupation Code |
15 |
|
Adopted Status |
Not Adopted |
|
T Stage |
T3 |
|
N Stage |
N3 |
|
6th Stage |
IIIC |
|
Differentiate |
Moderately differentiated |
|
Grade |
2 |
|
A Stage |
Regional |
|
Tumour Size |
41 |
|
Estrogen Status |
Positive |
|
Progesterone Status |
Positive |
|
Regional Node Examined |
5 |
|
Regional Node Positive |
1 |
Are you looking for expert help for your 7BUIS008W.1 Data Mining & Machine Learning Coursework? Get the best assignment help UK from our professional services, which will give you perfect guidance in your Computer Science assignments and Data Mining assignment help. In addition, we provide guidance with assignment examples specifically tailored to data mining and machine learning topics, so that you can learn and apply the concepts effectively. or personalized coursework help, our experts will deliver high-quality and plagiarism-free content for you. Take advantage of our service today to boost your grades!
Hire Assignment Helper Today!
Let's Book Your Work with Our Expert and Get High-Quality Content
