Each individual student should have his/her own labelled microfuge tube rack, kept in the dedicated freezer. These are available from the technician. Please remember to label all of your tubes appropriately with your initials. Unlabeled tubes or tubes stored in the wrong place will be disposed of without warning.
For the experiment you will need to use STERILE thin-walled PCR tubes (nuclease-free) and sterile pipette tips. When setting up your experiment it is critical to keep the reagents on ice and to replace them in the fridge as soon as possible.
You are provided with target/template DNA at 250 ng/ul (human placental DNA), your forward and reverse primers and enzyme mix, GreenTaq, containing Taq polymerase, reaction buffer, dNTPs and MgCl2.
You will need to set three reaction tubes, two replicates for each PCR reaction and one negative control reaction (i.e., without template DNA). First experiment you try 1 (ONE) reaction from below e.g. select Tm-5˚c and subsequent experiments select others.
For example:
2 reactions and 1 control at Tm-9˚c (9 degrees below melting temperature)
2 reactions and 1 control at Tm-7˚c
2 reactions and 1 control at Tm-5˚c
2 reactions and 1 control at Tm-3˚c
This will allow you to test a variety of temperatures and prove which is a good temperature by having data for that temperature as well as data for temperatures above and below it.
To do this you need to prepare a “master mix” – this will contain your primers primers, enzyme mix and nuclease free water needed for ALL your PCR reactions to be carried out at one time.
Instead of preparing exactly 3x the volume required for the 3 reaction tubes, you will prepare 3.5 x reaction mix volume, to avoid any shortage due to pipetting errors.
Your primers have been bought in from MWG Eurofins; the information sheets give the precise volumes of sterile water (NOT TBE Buffer) needed to give a concentration of 100 pmol/µl.
Pipette into each primer tube the correct volume (double check!) - as given in the data sheet – of DNase- and RNase-free water, and allow a couple of minutes for the primers to dissolve (creating your original stock solutions).
Then perform a dilution to produce a 10 pmol/µl working stock solution e.g. add 20 µl of original stock to 180 µl sterile DNase- and RNase-free water.
For each primer, the final concentration in the reaction will be 0.5 µM.
After use, your stock and working solutions should be kept frozen, in your personal rack, along with your other solutions.
Firstly, prepare the 3.5 x master mix in one tube: add the components as detailed below, then mix thoroughly (tap the tube, after mixing, drops of liquid stick to the top or sides, briefly (a few seconds) spin the tubes in the microfuge, ensuring it is balanced!!!).
The reason for preparing the master mix first is to facilitate accurate pipetting.
The Table below shows the additions needed to make the mastermix on the right (in bold), and the composition of the individual reaction tubes on the left.
The PCR Mix: Into your three, labelled reaction tubes, pipette the appropriate volume of master mix - 19 µl in this case.
Use a fresh pipette tip for each pipetting, and then discard!
Be very careful when transferring and mixing the reaction components, so that no kind of contamination occurs (tiny amounts of the wrong DNA might be amplified and give you the wrong bands). Your skin and hair will always have some bacteria on them and these bacteria contain DNases, therefore, if you touch your tubes with ungloved hands or touch your face with your glove and then continue with your work you will contaminate your reactions with DNase which will destroy all of your DNA and result in you not having any bands. You will have the same problem if you inadvertently spit into your tubes whilst talking over tubes with open lids.
Add an appropriate volume of DNA template solution (1 µl in this case) into each of the 2 experimental PCR tubes and the same volume of sterile deionised water to the control PCR tube.
When pipetting very small volumes, use the correct micropipette (e.g. 0-2 µl) – it can be helpful to touch the end of the pipette on the surface of the liquid you are pipetting into (e.g. master mix), then as you discharge, you see your 1 µl being taken up into the main body of liquid.
Mix the tube(s) gently and centrifuge briefly in a microfuge.
|
PCR |
Single reaction |
3.5x Master mix |
|
Forward primer (10pmol/µl) |
1 µl |
3.5 µl |
|
Reverse Primer (10pmol/µl) |
1 µl |
3.5 µl |
|
Taq polymerase mix |
10 µl |
35 µl |
|
Sterile deionized water |
7 µl |
24.5 µl |
|
Total reaction volume |
19 µl |
66.5 µl |
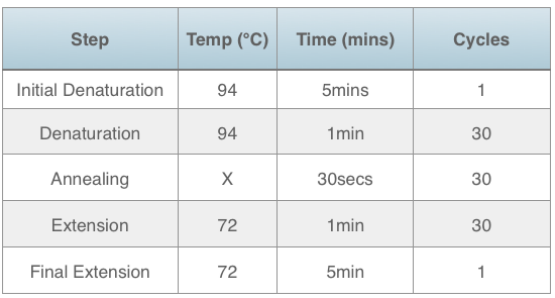
Insert the PCR tubes into the thermocycler.
Make sure that PCR machine is programmed as shown below (ask for help if needed, usually there is a program already available for the annealing temperature you want to use) and start the PCR program.
X = Tm of your primers minus 5°C./ selected Temperature (3°C , 7°C, 9°C)
The overall program should not usually last for more than 2 hours. We suggest you store/freeze half of each sample (labelled!) and analyze the other half (i.e. ~5-10 µl) by gel electrophoresis.
Buy Answer of This Assignment & Raise Your Grades
Request to Buy AnswerYou will prepare your own 1% (w/v) agarose gel, which needs to be dissolved in 0.5 x TBE (Tris-HCl, boric acid and EDTA). You can do this while your PCR is running.
by the particular gel tray you use).
Then, weigh the appropriate amount of agarose powder into a conical flask (3-4 times larger than gel volume e.g. if you want to prepare a 50 mL gel, you need a 200 mL flask).
Then add the appropriate amount of 0.5x TBE into the flask, swirl, and heat until boiling.
Take out the flask containing the hot liquid agarose solution (CARE! – it will be very hot - use heavy-duty gloves to avoid scalding !!) and place on the bench to cool.
Whilst the agarose is cooling, prepare your gel casting tray in such a way that the liquid agarose does not leak from the tray – this usually involves inserting the “comb”, which forms the sample slots (each slot needs to accommodate at least 10 µl of sample solution), and taping the ends of the tray with e.g. autoclave tape.
Note: Some gel systems are designed so that the gel tank can be used to seal the ends of the gel tray while the gel sets – so no tape is needed - The technicians can help guide you.
When the agarose solution becomes lukewarm, add 5µl of the DNA staining reagent GelRed. This is less than recommended by the manufacturer but is more than enough for this application.
Now pour your liquid agarose into the gel casting tray and wait for about 1 hour for the gel to set.
Load the gel in to the tank and immerse the gel in 0.5xTBE buffer (the same buffer used for making your gel). Note: the gel should be covered by 1-2mm of TBE buffer
Load your gel with 5µl of 100 bp DNA size markers (or whatever appropriate markers are available taking a note of what markers you used), and control and experimental samples (remember to use fresh tips for each sample!).
Once you have established the best temperature and you repeating your PCR reactions to obtain sufficient DNA for sequencing run only 5µl of your reaction on the gel.
Run the gel at a constant 90V to 100V – ensure that the sample slots are at the cathode/negative end of the tank, so that the DNA migrates towards the anode!!
When the marker dye (pre-added to the GreenTaq enzyme mix) has migrated down two-thirds of the gel (often less than 2 hours), turn off the current, and remove the gel from the electrophoresis tank.
Then visualize and record the DNA bands in a gel documentation system, take a photo, save this on the hard drive and email it to yourself. DO NOT USE A PEN/USB DRIVE. This should not take more than 10 min.
You can optimize your PCR experiment by altering the annealing temperature which can help when you don’t achieve a band or have multiple bands.
If you observe multiple bands, including your expected band, then you need to cut the appropriate band from the agarose gel with a sterile scalpel band and put the gel fragment into a sterile 1.5ml Eppendorf tube. This is done by visualising on a UV transluminator - you need to wear a UV eye protection shield.
Then you isolate the DNA fragment from the gel using a Qiagen gel extraction kit (protocol can be provided upon request). The purified DNA can then be sent for sequencing.
For this procedure, you will need to consult your supervisor.
Using your PCR product as a template for further PCR experiments
If, after gel electrophoresis, your band was very faint, this will mean that the amount of amplified DNA produced from the experiment is not enough for sequencing. Therefore you will need to “re-PCR” to increase the amount of DNA.
To do that you need to follow the same PCR procedure except: use 4 µl of amplified DNA as template DNA, and adjust the total volume with sterile DNase- and RNase-free water accordingly.
After re-PCR, check your amplified DNA by undertaking another gel electrophoresis analysis, following the same procedure as before.
For sequencing you need to send your amplified DNA and primers to MWG Eurofins. For each sequencing reaction the company needs 15 µl of amplified DNA at a concentration of 5 ng/µl for the forward sequencing reaction and the same again for the reverse sequencing reaction. They will also need 15 µl of 10 pmol/µl of each primer. Therefore, you need to quantify your purified DNA. You can do this by using spectrophotometric analysis.
To do this you will use the Biodrop, this works in the same way as a spectrophotometer but allows you to analyse just 2µl of your sample. Follow the instructions next to the Biodrop and ask a technician if you are unsure.
Briefly you will put 2µl of your ‘blank’ into the machine and press the ‘blank’ or ‘control’ button. You will wipe the ‘blank’ solution away gently but carefully with a small piece of tissue. You will then pipette 2µl of your reaction mixture into the machine and press the ‘reaction’ or ‘test’ button and make a note of the result. You will then carefully and gently wipe away your ‘test’ solution. Do not attempt to put the test solution back into your tube, it will now be contaminated with dust.
Your ‘blank’ or control will be 2ul of the EB buffer that you eluted your DNA in.
NOTE: mix ALL your successful amplified DNA samples together before removing your sample for absorbance measurement using the biodrop.
Use 1.5 ml Eppendorf tubes and label all the tubes clearly with a permanent fine marker pen using the following system:
For your DNA use your initialsD1 and initialsD2, for the primers use initialsF and initialsR ie for Jane Smith you would use JSD1, JSD2, JSF and JSR. If there are multiple students with the same initials we will ask you to use your username ie JS01 instead of just JS.
Once you have all your tubes you can now move on to the sequencing part of the project.
Small measurements
The “whatever” is an experimental unit of measurement used for teaching students how big things are (I may have just discovered it... Hoorah !).
If you chop a “whatever” (abbreviated to WH) into a thousand equal pieces, each will be a miliwhatever (mWH).
If you chop a mWH into a thousand equal pieces, each will be a microwhatever (µWH).
If you chop a µWH into a thousand equal pieces, each will be a nanowhatever (nWH).
If you chop a nWH into a thousand equal pieces, each will be a picowhatever (pWH).
This theory can be applied to volume (liters), mass (grammes), length (meters), volts (V), you name it !
The golden rule is to remember each unit is 1000 times smaller than it’s predecessor and the order the units come in, mili- then micro- then nano- then pico- and then femto-.
This is a bit like remembering the nomenclature describing the storage capacity of a computer hard disk, but in reverse (kilo-, mega-, giga-, tera- )! Funnily enough these designations can be applied to the whatever when talking about lots of them i.e. getting bigger rather than smaller.
Consequently you can have a thousand whatever they are equivalent to a kilowhatever (kWH) and a thousand kWH is equal to a megawhatever (mWH). If you were lucky (or rich) enough to collect a thousand MWH, you would have a gigawhatever (GWH) etc etc.
This nomenclature is used when describing the size of a DNA molecule however in this instance the elusive WH is replaced by the designation base pairs (bp). A plasmid would be (typically) 3-6 kbp (kilo base pairs) however it is not uncommon to measure genomic DNA in mega base pairs (Mbp) or even giga base pairs (Gbp).
The biodrop will calculate the concentration of your DNA for you, however, it is useful for you to understand how to do this if you used a cuvette and a standard spectrophotometer rather than the biodrop.
To calculate the amount of DNA, use the following formula:
Nominal DNA concentration (µg/ml) = OD260 x 50 x dilution factor. Here the dilution factor is 0 but if you use a cuvette rather than a biodrop or nanodrop you will have diluted your DNA in order to have sufficient volume.
DNA concentrations µM versus pmol/µl
You will often see concentrations for DNA written in µM, this is micro-molar.
Molarity is the concentration of a solution expressed as the number of moles per litre.
A 1 M solution is 1 mole in 1 litre, therefore, if you divide by 1000 you have µM instead of moles ie 1 µM is 1/1000th of a mole in a litre.
If you take 1 ml of this 1 µM solution then you have 1000 µl because a µl is 1/1000th of a ml. You also have 1000 pmol (pico-moles) because 1/1000th of a µM is a pmol.
Therefore: 1 µM = 1 pmol/µl
Achieve Higher Grades This Assignment & Raise Your Grades
Order Non Plagiarized AssignmentDo you need help on a Human Genome Project Laboratory Methods Revised? Then no need to worry! Our team of experts provides the online assignment help designed for students. We are here to assist you 24/7. We provide the best assignment writing services in fields like Human Resource Assignment Help, Science Assignment, Management etc… We also provide free assignment samples written by PhD expert writers. Contact us now and boost your academic grades!
Hire Assignment Helper Today!
Let's Book Your Work with Our Expert and Get High-Quality Content
